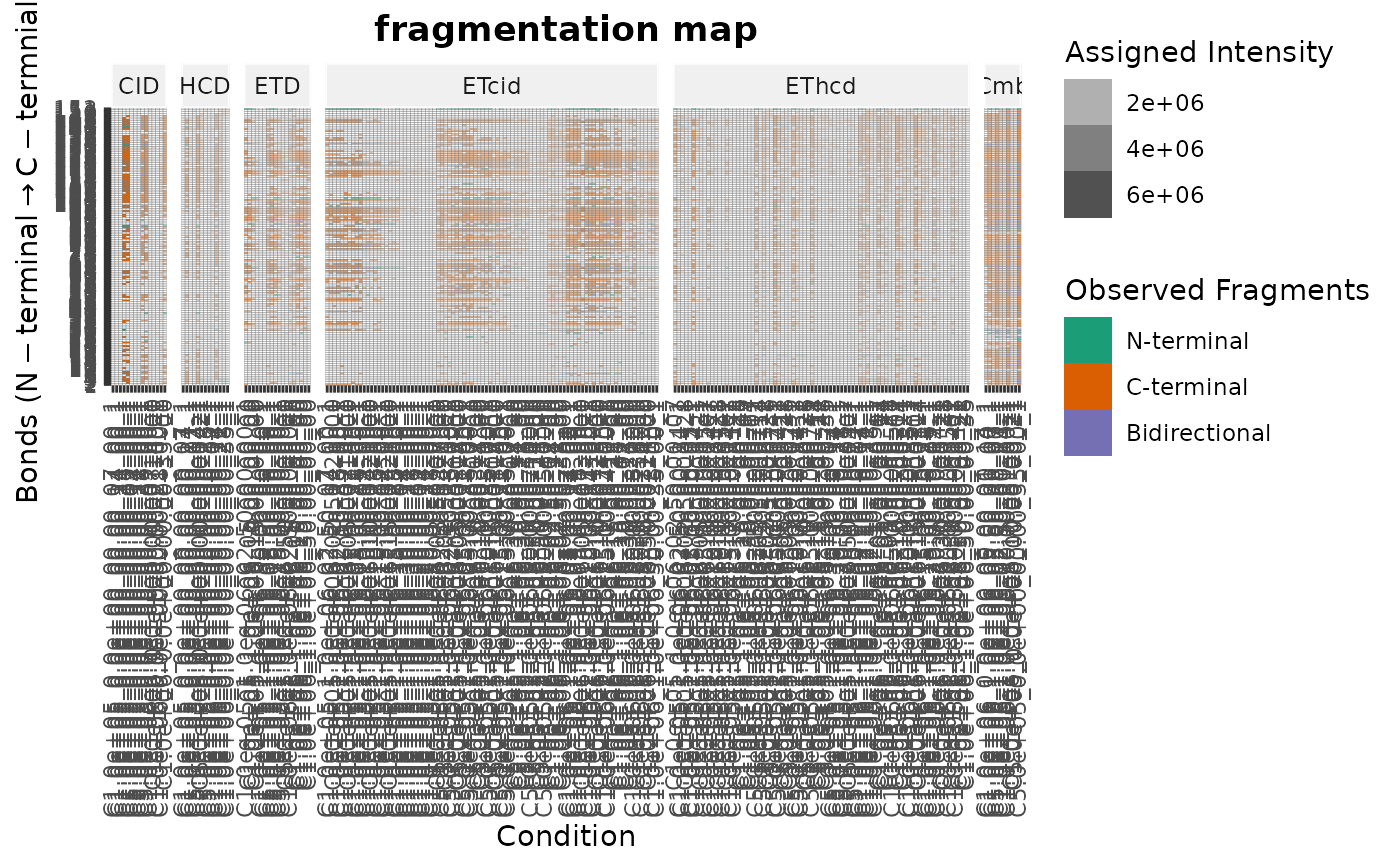
The NCBSet class is a container for a top-down proteomics experiment
similar to the TopDownSet
but instead of intensity values it just stores the information if a
bond is covered by a N-terminal (encoded as 1),
a C-terminal (encoded as 2)
and/or bidirectional fragments (encoded as 3).
# S4 method for NCBSet
bestConditions(
object,
n = ncol(object),
minN = 0L,
maximise = c("fragments", "bonds"),
...
)
# S4 method for NCBSet
fragmentationMap(
object,
nCombinations = 10,
cumCoverage = TRUE,
maximise = c("fragments", "bonds"),
labels = colnames(object),
alphaIntensity = TRUE,
...
)
# S4 method for NCBSet
show(object)
# S4 method for NCBSet
summary(object, what = c("conditions", "bonds"), ...)Arguments
| object |
|
|---|---|
| n |
|
| minN |
|
| maximise |
|
| nCombinations |
|
| cumCoverage |
|
| labels |
|
| alphaIntensity |
|
| what |
|
| ... | arguments passed to internal/other methods. added. |
Value
An NCBSet object.
Methods (by generic)
bestConditions: Best combination of conditions.Finds the best combination of conditions for highest coverage of fragments or bonds. If there are two (or more conditions) that would add the same number of fragments/bonds the one with the highest assigned intensity is used. Use
nto limit the number of iterations and combinations that should be returned. IfminNis set at leastminNfragments have to be added to the combinations. The function returns a 7-column matrix. The first column contains the index (Index) of the condition (column number). The next columns contain the newly added number of fragments or bonds (FragmentsAddedToCombination,BondsAddedToCombination), the fragments or bonds in the condition (FragmentsInCondition,BondsInCondition), and the cumulative coverage fragments or bonds (FragmentCoverage,BondCoverage).fragmentationMap: Plot fragmentation map.Plots a fragmentation map of the Protein. Use
nCombinationsto add another plot withnCombinationscombined conditions. IfcumCoverageisTRUE(default) these combinations increase the coverage cumulatively.summary: Summary statistics.Returns a
matrixwith some statistics: number of fragments, total/min/first quartile/median/mean/third quartile/maximum of intensity values.
Slots
rowViewsBiostrings::XStringViews, information about bonds (name, start, end, width, sequence), see Biostrings::XStringViews for details.
colDataS4Vectors::DataFrame, information about the MS2 experiments and the fragmentation conditions.
assayMatrix::dgCMatrix, coverage values of the bonds. The rows correspond to the bonds and the columns to the condition/run. It just stores values that are different from zero. If a bond is covered by an N-terminal fragment its encoded with
1, by an C-terminal fragmentl with2and by both fragment types/bidirectional by3respectively.filescharacter, files that were imported.processingcharacter, log messages.
See also
An
NCBSetis generated from an TopDownSet object.Biostrings::XStringViews for the row view interface.
Matrix::dgCMatrix for technical details about the coverage storage.
Author
Sebastian Gibb mail@sebastiangibb.de
Examples
## Example data
data(tds, package="topdownr")
## Aggregate technical replicates
tds <- aggregate(tds)
## Coercion into an NCBSet object
ncb <- as(tds, "NCBSet")
ncb
#> NCBSet object (0.34 Mb)
#> - - - Protein data - - -
#> Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
#> - - - Fragment data - - -
#> Number of N-terminal fragments: 438
#> Number of C-terminal fragments: 8039
#> Number of N- and C-terminal fragments: 140
#> - - - Condition data - - -
#> Number of conditions: 222
#> Number of scans: 222
#> Condition variables (60): File, Scan, ..., MedianIonInjectionTimeMs, AssignedIntensity
#> - - - Assay data - - -
#> Size of array: 152x222 (25.54% != 0)
#> - - - Processing information - - -
#> [2018-01-30 11:39:16] 14518 fragments [1949;351] matched (tolerance: 25 ppm, strategies ion/fragment: remove/remove).
#> [2018-01-30 11:39:16] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 222 conditions.
#> [2018-01-30 11:39:16] Recalculate median injection time based on: Mz, AgcTarget.
#> [2021-07-27 21:29:44] Aggregated 14518 fragments [1949;351] to 11026 fragments [1949;222].
#> [2021-07-27 21:29:45] Coerced TopDownSet into an NCBSet object; 8617 fragments [152;222].
head(summary(ncb))
#> Fragments Total Min Q1 Median Mean Q3 Max
#> C1.0e+05_1.0e+06_02.50_07_00_1 50 100 1 2 2 0.6578947 2 3
#> C1.0e+05_1.0e+06_02.50_14_00_1 48 93 1 2 2 0.6118421 2 2
#> C1.0e+05_1.0e+06_02.50_21_00 47 94 1 2 2 0.6184211 2 3
#> C1.0e+05_1.0e+06_02.50_28_00_1 55 108 1 2 2 0.7105263 2 2
#> C1.0e+05_1.0e+06_02.50_35_00_1 51 101 1 2 2 0.6644737 2 2
#> C1.0e+05_1.0e+06_02.50_00_07 63 124 1 2 2 0.8157895 2 2
# Accessing slots
rowViews(ncb)
#> Views on a 153-letter AAString subject
#> subject: GLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFT...PGDFGADAQGAMTKALELFRNDIAAKYKELGFQG
#> views:
#> start end width
#> [1] 1 1 1 [G]
#> [2] 1 2 2 [GL]
#> [3] 1 3 3 [GLS]
#> [4] 1 4 4 [GLSD]
#> [5] 1 5 5 [GLSDG]
#> ... ... ... ... ...
#> [148] 1 148 148 [GLSDGEWQQVLNVWGKVEADIAGHGQ...GADAQGAMTKALELFRNDIAAKYKE]
#> [149] 1 149 149 [GLSDGEWQQVLNVWGKVEADIAGHGQ...ADAQGAMTKALELFRNDIAAKYKEL]
#> [150] 1 150 150 [GLSDGEWQQVLNVWGKVEADIAGHGQ...DAQGAMTKALELFRNDIAAKYKELG]
#> [151] 1 151 151 [GLSDGEWQQVLNVWGKVEADIAGHGQ...AQGAMTKALELFRNDIAAKYKELGF]
#> [152] 1 152 152 [GLSDGEWQQVLNVWGKVEADIAGHGQ...QGAMTKALELFRNDIAAKYKELGFQ]
colData(ncb)
#> DataFrame with 222 rows and 60 columns
#> File Scan SpectrumIndex
#> <Rle> <integer> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 myo_1211_ETDReagentT.. 33 23.5
#> C1.0e+05_1.0e+06_02.50_14_00_1 myo_1211_ETDReagentT.. 36 26.5
#> C1.0e+05_1.0e+06_02.50_21_00 myo_1211_ETDReagentT.. 40 30.0
#> C1.0e+05_1.0e+06_02.50_28_00_1 myo_1211_ETDReagentT.. 43 33.5
#> C1.0e+05_1.0e+06_02.50_35_00_1 myo_1211_ETDReagentT.. 46 36.5
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 myo_1211_ETDReagentT.. 257 241.5
#> C1.0e+06_0.0e+00_00.00_00_14_1 myo_1211_ETDReagentT.. 260 244.5
#> C1.0e+06_0.0e+00_00.00_00_21_1 myo_1211_ETDReagentT.. 264 248.5
#> C1.0e+06_0.0e+00_00.00_00_28_1 myo_1211_ETDReagentT.. 268 252.5
#> C1.0e+06_0.0e+00_00.00_00_35_1 myo_1211_ETDReagentT.. 271 255.5
#> PeaksCount TotIonCurrent RetentionTime
#> <numeric> <numeric> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 134.0 14353074 380.439
#> C1.0e+05_1.0e+06_02.50_14_00_1 134.0 14563102 434.326
#> C1.0e+05_1.0e+06_02.50_21_00 148.0 14008273 497.289
#> C1.0e+05_1.0e+06_02.50_28_00_1 136.0 14285122 560.288
#> C1.0e+05_1.0e+06_02.50_35_00_1 134.5 14827734 614.126
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 50.0 731470 4641.38
#> C1.0e+06_0.0e+00_00.00_00_14_1 368.5 5725745 4690.68
#> C1.0e+06_0.0e+00_00.00_00_21_1 520.0 3477379 4756.43
#> C1.0e+06_0.0e+00_00.00_00_28_1 202.5 2015615 4822.08
#> C1.0e+06_0.0e+00_00.00_00_35_1 198.5 2889647 4870.88
#> BasePeakMz BasePeakIntensity.SpectraInformation
#> <numeric> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 16943 4867514
#> C1.0e+05_1.0e+06_02.50_14_00_1 16943 5007425
#> C1.0e+05_1.0e+06_02.50_21_00 16943 4605146
#> C1.0e+05_1.0e+06_02.50_28_00_1 16943 4835166
#> C1.0e+05_1.0e+06_02.50_35_00_1 16943 5029054
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 9065.461 31445.3
#> C1.0e+06_0.0e+00_00.00_00_14_1 16940.924 297301.0
#> C1.0e+06_0.0e+00_00.00_00_21_1 231.123 54766.1
#> C1.0e+06_0.0e+00_00.00_00_28_1 231.123 100368.1
#> C1.0e+06_0.0e+00_00.00_00_35_1 231.123 249724.3
#> LowMz HighMz PrecursorMz
#> <numeric> <numeric> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 762.391 17146.1 1211.7
#> C1.0e+05_1.0e+06_02.50_14_00_1 762.391 17146.1 1211.7
#> C1.0e+05_1.0e+06_02.50_21_00 762.391 17146.1 1211.7
#> C1.0e+05_1.0e+06_02.50_28_00_1 762.391 17146.6 1211.7
#> C1.0e+05_1.0e+06_02.50_35_00_1 762.390 17146.1 1211.7
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 1146.114 16941.01 1211.72
#> C1.0e+06_0.0e+00_00.00_00_14_1 244.634 16940.92 1211.72
#> C1.0e+06_0.0e+00_00.00_00_21_1 200.139 5468.80 1211.72
#> C1.0e+06_0.0e+00_00.00_00_28_1 200.139 1406.28 1211.72
#> C1.0e+06_0.0e+00_00.00_00_35_1 200.139 1049.08 1211.72
#> SpectrumId Condition
#> <character> <integer>
#> C1.0e+05_1.0e+06_02.50_07_00_1 controllerType=0 con.. 12
#> C1.0e+05_1.0e+06_02.50_14_00_1 controllerType=0 con.. 14
#> C1.0e+05_1.0e+06_02.50_21_00 controllerType=0 con.. 16
#> C1.0e+05_1.0e+06_02.50_28_00_1 controllerType=0 con.. 18
#> C1.0e+05_1.0e+06_02.50_35_00_1 controllerType=0 con.. 20
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 controllerType=0 con.. 154
#> C1.0e+06_0.0e+00_00.00_00_14_1 controllerType=0 con.. 156
#> C1.0e+06_0.0e+00_00.00_00_21_1 controllerType=0 con.. 158
#> C1.0e+06_0.0e+00_00.00_00_28_1 controllerType=0 con.. 160
#> C1.0e+06_0.0e+00_00.00_00_35_1 controllerType=0 con.. 162
#> FilterString.HeaderInformation
#> <character>
#> C1.0e+05_1.0e+06_02.50_07_00_1 FTMS + p NSI sa Full..
#> C1.0e+05_1.0e+06_02.50_14_00_1 FTMS + p NSI sa Full..
#> C1.0e+05_1.0e+06_02.50_21_00 FTMS + p NSI sa Full..
#> C1.0e+05_1.0e+06_02.50_28_00_1 FTMS + p NSI sa Full..
#> C1.0e+05_1.0e+06_02.50_35_00_1 FTMS + p NSI sa Full..
#> ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 FTMS + p NSI Full ms..
#> C1.0e+06_0.0e+00_00.00_00_14_1 FTMS + p NSI Full ms..
#> C1.0e+06_0.0e+00_00.00_00_21_1 FTMS + p NSI Full ms..
#> C1.0e+06_0.0e+00_00.00_00_28_1 FTMS + p NSI Full ms..
#> C1.0e+06_0.0e+00_00.00_00_35_1 FTMS + p NSI Full ms..
#> SupplementalActivation RtMin Tic
#> <Rle> <numeric> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 TRUE 6.34064 165522520
#> C1.0e+05_1.0e+06_02.50_14_00_1 TRUE 7.23878 168781032
#> C1.0e+05_1.0e+06_02.50_21_00 TRUE 8.28815 159144752
#> C1.0e+05_1.0e+06_02.50_28_00_1 TRUE 9.33814 164578728
#> C1.0e+05_1.0e+06_02.50_35_00_1 TRUE 10.23544 169997320
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 FALSE 77.3563 46251106
#> C1.0e+06_0.0e+00_00.00_00_14_1 FALSE 78.1780 36784694
#> C1.0e+06_0.0e+00_00.00_00_21_1 FALSE 79.2739 6848908
#> C1.0e+06_0.0e+00_00.00_00_28_1 FALSE 80.3680 4623504
#> C1.0e+06_0.0e+00_00.00_00_35_1 FALSE 81.1814 6598797
#> BasePeakPosition
#> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 1304.93
#> C1.0e+05_1.0e+06_02.50_14_00_1 1304.93
#> C1.0e+05_1.0e+06_02.50_21_00 1304.93
#> C1.0e+05_1.0e+06_02.50_28_00_1 1304.93
#> C1.0e+05_1.0e+06_02.50_35_00_1 1304.89
#> ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 1211.746
#> C1.0e+06_0.0e+00_00.00_00_14_1 1211.755
#> C1.0e+06_0.0e+00_00.00_00_21_1 231.123
#> C1.0e+06_0.0e+00_00.00_00_28_1 195.107
#> C1.0e+06_0.0e+00_00.00_00_35_1 231.123
#> BasePeakIntensity.HeaderInformation
#> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 3773813
#> C1.0e+05_1.0e+06_02.50_14_00_1 3854662
#> C1.0e+05_1.0e+06_02.50_21_00 3627429
#> C1.0e+05_1.0e+06_02.50_28_00_1 3772335
#> C1.0e+05_1.0e+06_02.50_35_00_1 3699570
#> ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 30933570.0
#> C1.0e+06_0.0e+00_00.00_00_14_1 745855.8
#> C1.0e+06_0.0e+00_00.00_00_21_1 49163.1
#> C1.0e+06_0.0e+00_00.00_00_28_1 99657.5
#> C1.0e+06_0.0e+00_00.00_00_35_1 222407.6
#> IonInjectionTimeMs ElapsedScanTimeSec HcdEnergy
#> <numeric> <numeric> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 4.7190 17.8500 NaN
#> C1.0e+05_1.0e+06_02.50_14_00_1 4.4465 17.8405 NaN
#> C1.0e+05_1.0e+06_02.50_21_00 5.6220 17.8870 NaN
#> C1.0e+05_1.0e+06_02.50_28_00_1 4.9790 17.8605 NaN
#> C1.0e+05_1.0e+06_02.50_35_00_1 4.2125 17.8315 NaN
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 41.5310 16.1035 7
#> C1.0e+06_0.0e+00_00.00_00_14_1 36.3915 15.8980 14
#> C1.0e+06_0.0e+00_00.00_00_21_1 38.7175 15.9905 21
#> C1.0e+06_0.0e+00_00.00_00_28_1 33.2765 15.7745 28
#> C1.0e+06_0.0e+00_00.00_00_35_1 34.1280 15.8080 35
#> ConversionParameterB ConversionParameterC
#> <numeric> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 211703103 -146989184
#> C1.0e+05_1.0e+06_02.50_14_00_1 211703103 -146989183
#> C1.0e+05_1.0e+06_02.50_21_00 211703102 -146989182
#> C1.0e+05_1.0e+06_02.50_28_00_1 211703101 -146989181
#> C1.0e+05_1.0e+06_02.50_35_00_1 211703101 -146989180
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 211702383 -146988183
#> C1.0e+06_0.0e+00_00.00_00_14_1 211702382 -146988182
#> C1.0e+06_0.0e+00_00.00_00_21_1 211702381 -146988181
#> C1.0e+06_0.0e+00_00.00_00_28_1 211702381 -146988180
#> C1.0e+06_0.0e+00_00.00_00_35_1 211702380 -146988179
#> TemperatureCompPpm SpaceChargeCompPpm RawOvFtt
#> <Rle> <Rle> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 -14.44 -0.18 778212
#> C1.0e+05_1.0e+06_02.50_14_00_1 -14.44 -0.18 746397
#> C1.0e+05_1.0e+06_02.50_21_00 -14.44 -0.18 897244
#> C1.0e+05_1.0e+06_02.50_28_00_1 -14.44 -0.18 810213
#> C1.0e+05_1.0e+06_02.50_35_00_1 -14.44 -0.18 720653
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 -14.56 -1.76 1884474
#> C1.0e+06_0.0e+00_00.00_00_14_1 -14.56 -1.76 1354524
#> C1.0e+06_0.0e+00_00.00_00_21_1 -14.56 -1.76 275211
#> C1.0e+06_0.0e+00_00.00_00_28_1 -14.56 -1.76 158895
#> C1.0e+06_0.0e+00_00.00_00_35_1 -14.56 -1.76 229971
#> InjectionT0 NrCentroids SelectedMass1
#> <numeric> <numeric> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 0 3283.0 1211.7
#> C1.0e+05_1.0e+06_02.50_14_00_1 0 3428.5 1211.7
#> C1.0e+05_1.0e+06_02.50_21_00 0 3629.0 1211.7
#> C1.0e+05_1.0e+06_02.50_28_00_1 0 3464.5 1211.7
#> C1.0e+05_1.0e+06_02.50_35_00_1 0 3341.5 1211.7
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 0.000 3128.0 1211.72
#> C1.0e+06_0.0e+00_00.00_00_14_1 -0.048 6190.5 1211.72
#> C1.0e+06_0.0e+00_00.00_00_21_1 -0.055 4913.0 1211.72
#> C1.0e+06_0.0e+00_00.00_00_28_1 0.033 2346.0 1211.72
#> C1.0e+06_0.0e+00_00.00_00_35_1 0.035 2365.5 1211.72
#> Activation1 Energy1 SelectedMass2 Activation2
#> <Rle> <Rle> <Rle> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 ETD 2.5 0 CID
#> C1.0e+05_1.0e+06_02.50_14_00_1 ETD 2.5 0 CID
#> C1.0e+05_1.0e+06_02.50_21_00 ETD 2.5 0 CID
#> C1.0e+05_1.0e+06_02.50_28_00_1 ETD 2.5 0 CID
#> C1.0e+05_1.0e+06_02.50_35_00_1 ETD 2.5 0 CID
#> ... ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 HCD 7 NaN NA
#> C1.0e+06_0.0e+00_00.00_00_14_1 HCD 14 NaN NA
#> C1.0e+06_0.0e+00_00.00_00_21_1 HCD 21 NaN NA
#> C1.0e+06_0.0e+00_00.00_00_28_1 HCD 28 NaN NA
#> C1.0e+06_0.0e+00_00.00_00_35_1 HCD 35 NaN NA
#> Energy2 EtdActivation CidActivation
#> <numeric> <Rle> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 7 2.5 7
#> C1.0e+05_1.0e+06_02.50_14_00_1 14 2.5 14
#> C1.0e+05_1.0e+06_02.50_21_00 21 2.5 21
#> C1.0e+05_1.0e+06_02.50_28_00_1 28 2.5 28
#> C1.0e+05_1.0e+06_02.50_35_00_1 35 2.5 35
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 NaN NaN NaN
#> C1.0e+06_0.0e+00_00.00_00_14_1 NaN NaN NaN
#> C1.0e+06_0.0e+00_00.00_00_21_1 NaN NaN NaN
#> C1.0e+06_0.0e+00_00.00_00_28_1 NaN NaN NaN
#> C1.0e+06_0.0e+00_00.00_00_35_1 NaN NaN NaN
#> HcdActivation Activation StartTime EndTime
#> <numeric> <Rle> <numeric> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 NaN ETcid 6.01 6.5
#> C1.0e+05_1.0e+06_02.50_14_00_1 NaN ETcid 7.01 7.5
#> C1.0e+05_1.0e+06_02.50_21_00 NaN ETcid 8.01 8.5
#> C1.0e+05_1.0e+06_02.50_28_00_1 NaN ETcid 9.01 9.5
#> C1.0e+05_1.0e+06_02.50_35_00_1 NaN ETcid 10.01 10.5
#> ... ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 7 HCD 77.01 77.5
#> C1.0e+06_0.0e+00_00.00_00_14_1 14 HCD 78.01 78.5
#> C1.0e+06_0.0e+00_00.00_00_21_1 21 HCD 79.01 79.5
#> C1.0e+06_0.0e+00_00.00_00_28_1 28 HCD 80.01 80.5
#> C1.0e+06_0.0e+00_00.00_00_35_1 35 HCD 81.01 81.5
#> ActivationType Ce SteppedCe SteppedCeValue
#> <Rle> <Rle> <Rle> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 ETD NaN NaN
#> C1.0e+05_1.0e+06_02.50_14_00_1 ETD NaN NaN
#> C1.0e+05_1.0e+06_02.50_21_00 ETD NaN NaN
#> C1.0e+05_1.0e+06_02.50_28_00_1 ETD NaN NaN
#> C1.0e+05_1.0e+06_02.50_35_00_1 ETD NaN NaN
#> ... ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 HCD 25 False 5
#> C1.0e+06_0.0e+00_00.00_00_14_1 HCD 25 False 5
#> C1.0e+06_0.0e+00_00.00_00_21_1 HCD 25 False 5
#> C1.0e+06_0.0e+00_00.00_00_28_1 HCD 25 False 5
#> C1.0e+06_0.0e+00_00.00_00_35_1 HCD 25 False 5
#> MultistageActivation Ethcd
#> <Rle> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 FALSE
#> C1.0e+05_1.0e+06_02.50_14_00_1 FALSE
#> C1.0e+05_1.0e+06_02.50_21_00 FALSE
#> C1.0e+05_1.0e+06_02.50_28_00_1 FALSE
#> C1.0e+05_1.0e+06_02.50_35_00_1 FALSE
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 FALSE
#> C1.0e+06_0.0e+00_00.00_00_14_1 FALSE
#> C1.0e+06_0.0e+00_00.00_00_21_1 FALSE
#> C1.0e+06_0.0e+00_00.00_00_28_1 FALSE
#> C1.0e+06_0.0e+00_00.00_00_35_1 FALSE
#> UseCalibratedEtdParameters EtdReactionTime
#> <Rle> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 False 2.5
#> C1.0e+05_1.0e+06_02.50_14_00_1 False 2.5
#> C1.0e+05_1.0e+06_02.50_21_00 False 2.5
#> C1.0e+05_1.0e+06_02.50_28_00_1 False 2.5
#> C1.0e+05_1.0e+06_02.50_35_00_1 False 2.5
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_14_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_21_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_28_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_35_1 NaN
#> EtdReagentTarget EtdSupplementalActivation
#> <Rle> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 1e+06 True
#> C1.0e+05_1.0e+06_02.50_14_00_1 1e+06 True
#> C1.0e+05_1.0e+06_02.50_21_00 1e+06 True
#> C1.0e+05_1.0e+06_02.50_28_00_1 1e+06 True
#> C1.0e+05_1.0e+06_02.50_35_00_1 1e+06 True
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_14_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_21_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_28_1 NaN
#> C1.0e+06_0.0e+00_00.00_00_35_1 NaN
#> SupplementalActivationCe AgcTarget Activationq
#> <numeric> <Rle> <Rle>
#> C1.0e+05_1.0e+06_02.50_07_00_1 7 1e+05 NaN
#> C1.0e+05_1.0e+06_02.50_14_00_1 14 1e+05 NaN
#> C1.0e+05_1.0e+06_02.50_21_00 21 1e+05 NaN
#> C1.0e+05_1.0e+06_02.50_28_00_1 28 1e+05 NaN
#> C1.0e+05_1.0e+06_02.50_35_00_1 35 1e+05 NaN
#> ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 NaN 1e+06 NaN
#> C1.0e+06_0.0e+00_00.00_00_14_1 NaN 1e+06 NaN
#> C1.0e+06_0.0e+00_00.00_00_21_1 NaN 1e+06 NaN
#> C1.0e+06_0.0e+00_00.00_00_28_1 NaN 1e+06 NaN
#> C1.0e+06_0.0e+00_00.00_00_35_1 NaN 1e+06 NaN
#> TargetedMassList Mz Charge Sample
#> <character> <Rle> <Rle> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 (mz=1211.7012 z=1 na.. 1211.7 14 1
#> C1.0e+05_1.0e+06_02.50_14_00_1 (mz=1211.7014 z=1 na.. 1211.7 14 2
#> C1.0e+05_1.0e+06_02.50_21_00 (mz=1211.7016 z=1 na.. 1211.7 14 3
#> C1.0e+05_1.0e+06_02.50_28_00_1 (mz=1211.7018 z=1 na.. 1211.7 14 4
#> C1.0e+05_1.0e+06_02.50_35_00_1 (mz=1211.702 z=1 nam.. 1211.7 14 5
#> ... ... ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 (mz=1211.7154 z=1 na.. 1211.7 14 218
#> C1.0e+06_0.0e+00_00.00_00_14_1 (mz=1211.7156 z=1 na.. 1211.7 14 219
#> C1.0e+06_0.0e+00_00.00_00_21_1 (mz=1211.7158 z=1 na.. 1211.7 14 220
#> C1.0e+06_0.0e+00_00.00_00_28_1 (mz=1211.716 z=1 nam.. 1211.7 14 221
#> C1.0e+06_0.0e+00_00.00_00_35_1 (mz=1211.7162 z=1 na.. 1211.7 14 222
#> MedianIonInjectionTimeMs AssignedIntensity
#> <Rle> <numeric>
#> C1.0e+05_1.0e+06_02.50_07_00_1 4.837 3584559
#> C1.0e+05_1.0e+06_02.50_14_00_1 4.837 3693918
#> C1.0e+05_1.0e+06_02.50_21_00 4.837 3590927
#> C1.0e+05_1.0e+06_02.50_28_00_1 4.837 3935368
#> C1.0e+05_1.0e+06_02.50_35_00_1 4.837 3773703
#> ... ... ...
#> C1.0e+06_0.0e+00_00.00_00_07_1 43.14 105927.6
#> C1.0e+06_0.0e+00_00.00_00_14_1 43.14 1786586.1
#> C1.0e+06_0.0e+00_00.00_00_21_1 43.14 400658.5
#> C1.0e+06_0.0e+00_00.00_00_28_1 43.14 53933.0
#> C1.0e+06_0.0e+00_00.00_00_35_1 43.14 56438.2
head(assayData(ncb))
#> 6 x 222 sparse Matrix of class "dgCMatrix"
#> [[ suppressing 222 column names ‘C1.0e+05_1.0e+06_02.50_07_00_1’, ‘C1.0e+05_1.0e+06_02.50_14_00_1’, ‘C1.0e+05_1.0e+06_02.50_21_00’ ... ]]
#>
#> bond001 2 2 2 2 2 2 2 . . . 2 2 2 . 2 2 2 2 . . . . . . . . 2 . . . . . . . . .
#> bond002 . . . . . 2 2 2 . . . . . 2 . . 2 2 2 . . . . . . . 2 . . . . . . . . .
#> bond003 . . . . . 2 . . . . . . . . . . 2 . . . . . . . . . . . . . . . . . . .
#> bond004 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
#> bond005 . . . . . 2 . . . . . . . . . . 2 . . . . . . . . . . . . . . . . . . .
#> bond006 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
#>
#> bond001 . 2 . . . . . . . . . . . . . . . . . . . . . . . 2 2 2 . . 2 . . . . 2
#> bond002 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 . 2 . . . .
#> bond003 . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 2 . 2 . . . 2
#> bond004 . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 2 . 2 . . . .
#> bond005 . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 . . 2 . . . .
#> bond006 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 . 2 . . . .
#>
#> bond001 2 2 2 2 2 . 2 . . 2 2 . 2 . 2 2 2 . . . . . . . . . 2 2 . . . . . . . .
#> bond002 . . 2 . 2 . 2 . 1 . 2 . 2 2 2 2 2 2 . 1 2 . . . . 2 2 2 . . . 2 . . . 2
#> bond003 . 2 . . 2 . 2 1 1 . . . . . . 2 2 . . 1 . . . . . . 2 . . . . . . . . .
#> bond004 . . . . . . . . 1 . . . . . . . . . . . . . . . . . . 2 . . . . . . . .
#> bond005 . . . . 2 . . . . . . . . . . 2 . . . . . . . . . . . . . . . . . . . .
#> bond006 . . . . 2 . 2 . . . . . . . . . . 2 . . . . . . . . . . . . . . . . . .
#>
#> bond001 . 2 . . . . . . . . . 2 . . . . . . . . . . . . . . . . 2 2 2 . . 2 . .
#> bond002 . 2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 . 2 2 1
#> bond003 . 2 . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 . 2 . . 2 1
#> bond004 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 2 . 2 1
#> bond005 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 2 . 2 .
#> bond006 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 .
#>
#> bond001 . . 2 2 2 2 . 2 2 . . . 2 2 2 2 2 2 2 2 . . . 2 . . 2 . . 2 2 2 . . . .
#> bond002 1 1 2 . . 2 . 2 2 2 . 1 2 2 2 2 2 . 2 2 2 . 1 2 2 . 2 . 2 2 2 2 . . . .
#> bond003 1 1 . . . 2 . 2 2 2 1 1 . . . . . . 2 2 2 1 1 2 . . . . . 2 . . . 1 . .
#> bond004 . . . . . . . 2 2 2 1 1 . . . . . . 2 2 2 . . . . . . . . . . . . . . .
#> bond005 . . . . . . . 2 2 . . . . . . . . . 2 2 . . . . . . . . . 2 . . . 1 . .
#> bond006 . . . . . . . . . . . . . . 2 . 2 . . . . . . . . . . . . 2 . . . . . .
#>
#> bond001 2 . . . 2 2 . 2 . . . . . . . 2 2 . . . . . . . . . . . . . . . 2 . 2 2
#> bond002 . . 2 . 2 2 2 2 . . . . . 2 . . 2 . . . . . . . . . . . . . . . 2 . . 2
#> bond003 . . . . 2 2 . . 1 2 . . . . . . . . . 1 . . . . . . . . . . . . . . . .
#> bond004 . . . . . 2 . 1 . . . . . . . . 2 . 1 . . . . . . . . . . . . . . . . 2
#> bond005 . . . . 2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
#> bond006 . . . . . 2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
#>
#> bond001 . 2 . . . .
#> bond002 2 2 . . 1 1
#> bond003 . . . 1 1 1
#> bond004 2 . 2 1 1 1
#> bond005 . . 2 . . .
#> bond006 . . 2 . . .
# Accessing colData
ncb$Mz
#> numeric-Rle of length 222 with 1 run
#> Lengths: 222
#> Values : 1211.7
# Subsetting
# First 100 bonds
ncb[1:100]
#> NCBSet object (0.28 Mb)
#> - - - Protein data - - -
#> Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
#> - - - Fragment data - - -
#> Number of N-terminal fragments: 263
#> Number of C-terminal fragments: 4191
#> Number of N- and C-terminal fragments: 120
#> - - - Condition data - - -
#> Number of conditions: 222
#> Number of scans: 222
#> Condition variables (60): File, Scan, ..., MedianIonInjectionTimeMs, AssignedIntensity
#> - - - Assay data - - -
#> Size of array: 100x222 (20.60% != 0)
#> - - - Processing information - - -
#> [2018-01-30 11:39:16] 14518 fragments [1949;351] matched (tolerance: 25 ppm, strategies ion/fragment: remove/remove).
#> [2018-01-30 11:39:16] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 222 conditions.
#> [2018-01-30 11:39:16] Recalculate median injection time based on: Mz, AgcTarget.
#> [2021-07-27 21:29:44] Aggregated 14518 fragments [1949;351] to 11026 fragments [1949;222].
#> [2021-07-27 21:29:45] Coerced TopDownSet into an NCBSet object; 8617 fragments [152;222].
#> [2021-07-27 21:29:45] Subsetted 8617 fragments [152;222] to 4574 fragments [100;222].
# Just bond 152
ncb["bond152"]
#> NCBSet object (0.22 Mb)
#> - - - Protein data - - -
#> Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
#> - - - Fragment data - - -
#> Number of N-terminal fragments: 97
#> Number of C-terminal fragments: 0
#> Number of N- and C-terminal fragments: 0
#> - - - Condition data - - -
#> Number of conditions: 222
#> Number of scans: 222
#> Condition variables (60): File, Scan, ..., MedianIonInjectionTimeMs, AssignedIntensity
#> - - - Assay data - - -
#> Size of array: 1x222 (43.69% != 0)
#> - - - Processing information - - -
#> [2018-01-30 11:39:16] 14518 fragments [1949;351] matched (tolerance: 25 ppm, strategies ion/fragment: remove/remove).
#> [2018-01-30 11:39:16] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 222 conditions.
#> [2018-01-30 11:39:16] Recalculate median injection time based on: Mz, AgcTarget.
#> [2021-07-27 21:29:44] Aggregated 14518 fragments [1949;351] to 11026 fragments [1949;222].
#> [2021-07-27 21:29:45] Coerced TopDownSet into an NCBSet object; 8617 fragments [152;222].
#> [2021-07-27 21:29:45] Subsetted 8617 fragments [152;222] to 97 fragments [1;222].
# Condition 1 to 10
ncb[, 1:10]
#> NCBSet object (0.08 Mb)
#> - - - Protein data - - -
#> Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
#> - - - Fragment data - - -
#> Number of N-terminal fragments: 11
#> Number of C-terminal fragments: 372
#> Number of N- and C-terminal fragments: 2
#> - - - Condition data - - -
#> Number of conditions: 10
#> Number of scans: 10
#> Condition variables (60): File, Scan, ..., MedianIonInjectionTimeMs, AssignedIntensity
#> - - - Assay data - - -
#> Size of array: 152x10 (25.33% != 0)
#> - - - Processing information - - -
#> [2018-01-30 11:39:16] 14518 fragments [1949;351] matched (tolerance: 25 ppm, strategies ion/fragment: remove/remove).
#> [2018-01-30 11:39:16] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 222 conditions.
#> [2018-01-30 11:39:16] Recalculate median injection time based on: Mz, AgcTarget.
#> [2021-07-27 21:29:44] Aggregated 14518 fragments [1949;351] to 11026 fragments [1949;222].
#> [2021-07-27 21:29:45] Coerced TopDownSet into an NCBSet object; 8617 fragments [152;222].
#> [2021-07-27 21:29:45] Subsetted 8617 fragments [152;222] to 385 fragments [152;10].
# Plot fragmentation map
fragmentationMap(ncb)
#> Warning: Raster pixels are placed at uneven vertical intervals and will be shifted. Consider using geom_tile() instead.